12.1. Importing data from ROOT files¶
Importing data from ROOT files is easy: Points and Histogram/Histogram2d objects may be constructed directly from
TMatrixD/TMatrixF/TH1/TH2 objects. An example may be found in the following file
misc_importing_from_root.py.
Here we will discuss only main functions and points. Firstly let us create TH1D, TH2D and TMatrixD.
# Create ROOT objects
roothist1d = R.TH1D('testhist', 'testhist', 10, -5, 5)
roothist2d = R.TH2D('testhist', 'testhist', 20, 0, 10, 24, 0, 12)
Then, after ROOT objects are filled, or just read from the ROOT file, the GNA objects may be created with their regular constructors:
# Create Points
p1 = C.Points(roothist1d)
p2 = C.Points(roothist2d)
# Create Histograms
h1d = C.Histogram(roothist1d)
The single precision versions TH1F, TH2F and TMatrixF are valid as well.
- It should be noted, that
Points loose any information on bin edges.
Internally TH2 keeps the data in transposed order, i.e. first index represents y-coordinate, while second index represents x-coordinate. When converted to Histogram2d the memory order is changed implicitly to x/y for first/second index.
The difference between array-like and histogram-like representation may be seen from the following figures.
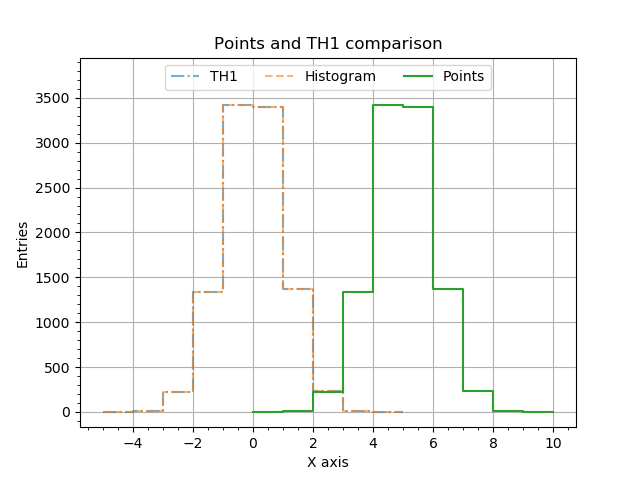
1d histograms plotted from ROOT.TH1D, GNA.Histogram and GNA.Points.¶
When Points object is plotted as histogram, it is assumed that first bin starts at 0 and bin width is 1.
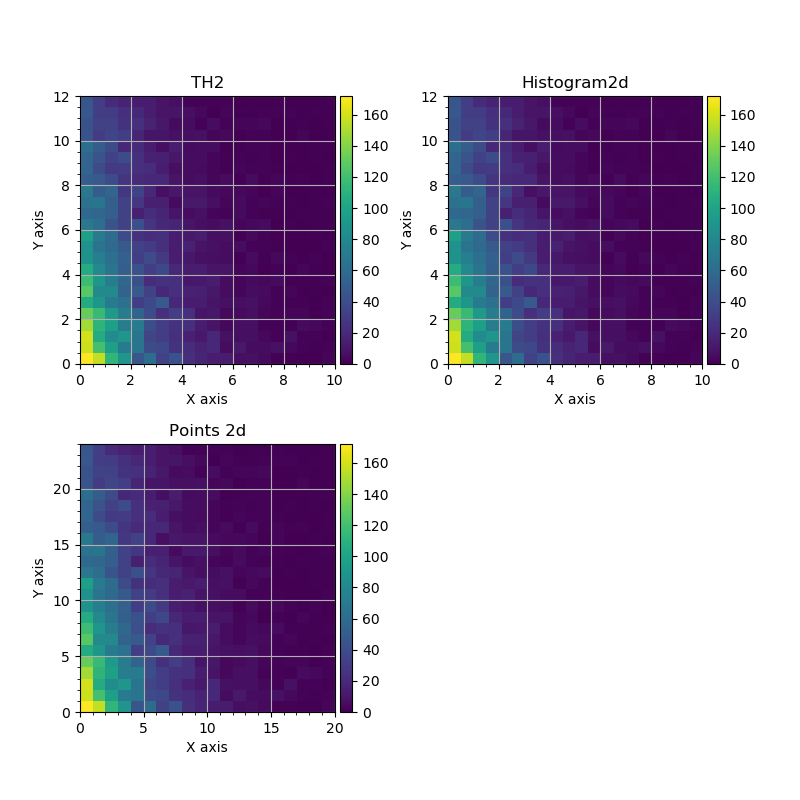
2d histograms plotted from ROOT.TH2D, GNA.Histogram2d and GNA.Points.¶
The same applies to Points object is plotted as 2d histogram.